Calibrate 3D Super Resolution Module
Follow the steps below to ensure the 3D Super Resolution Module is fully calibrated before acquiring a 3D SMLM protocol.
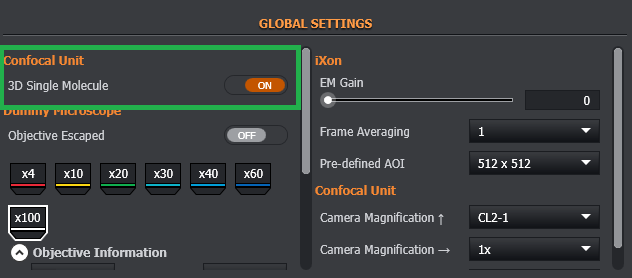
Engage 3D Super Resolution Module by switching on the 3D Single Molecule option which can be found by default in confocal unit global settings in Channel Manager. If you cannot find it here, go to Assign Feature Location to see where the feature is located.
Acquire the calibration stack.
Use a bead sample, and beads of 100 to 200 nm size
Engage the 3D Super Resolution Module (3D Single Molecule ON)
Focus your beads and set the centre of the bead sample
Set a Z stack with 10 nm Z step and to cover a 1,2 um range
Acquire the calibration stack over a small ROI window (512x512, or 256 x256)
Ensure that within the acquisition window, you have 5-10 beads well separated that do not overlap each other
Acquire the Z stack.
The user should see the distortion of the beads horizontally and vertically as the focus moves through Z.
After the acquisition, the calibration of the 3D super-resolution module to retrieve the super resolved axial information can be done in Picasso.
Calibrate the axial (Z) encoding in Picasso.
Adjust the parameters dependent on the camera and objective used (see “Andor Cameras Parameters” file).
Calibrate the 3D Super Resolution Module (3D -> Calibrate 3D).
Enter the step size used to acquire the calibration file.
Analyse and validate the calibration.